Co-leader: Zachary Morris, MD, PhD
Associate Professor and Chair
Department of Human Oncology
University of Wisconsin

Co-leader: Jamey Weichert, PhD
Professor
Department of Radiology
University of Wisconsin

Co-leader: Paul Harari, MD
Professor
Department of Human Oncology
University of Wisconsin
Summary:
Despite advances in treatments including the introduction of immunotherapies, metastatic or recurrent head and neck squamous cell cancers are typically incurable and, to begin changing this, we propose to develop and investigate a new treatment approach that combines the most common form of immunotherapy (immune checkpoint inhibitors) with targeted radionuclide therapy. Radiation can damage tumors in a way that renders them more responsive to immunotherapies, however conventional radiation therapy typically cannot treat all tumor sites in patients with metastatic disease because of toxicities and the inability to target small tumors that are not identified on imaging scans. Targeted radionuclide therapies go throughout the body to preferentially deliver radiation to tumors wherever they are located. In this University of Wisconsin Head and Neck SPORE grant project we will (1) determine the effect of a novel targeted radionuclide therapy on the susceptibility of head and neck squamous cell carcinoma tumor models to immune response; and (2) test the effectiveness of this type of treatment when combined with immune checkpoint blockade in order to determine which combinations are safest and most beneficial in patients with metastatic or recurrent head and neck cancer. The insights and treatment regimens developed in these studies should enable translation to advanced phase clinical testing in patients with head and neck cancers and may guide the development of similar combinations of targeted radionuclide therapies and immunotherapies for other types of metastatic cancer
Specific Aims:
-
Aim 1. Evaluate the capacity of 90Y-, 177Lu-, and 225Ac-NM600 to deliver immunomodulatory radiation selectively to syngeneic murine HNSCC tumor models.
-
Aim 2. Compare the ability of 90Y-, 177Lu-, and 225Ac-NM600 to augment anti-tumor immune response when delivered with anti-PD-1 in syngeneic murine HNSCC tumor models.
-
Aim 3. Perform a phase I trial to evaluate the safety of combining NM600 TRT and anti-PD1 therapy in patients with metastatic/recurrent HNSCC.
Collaborators:

Bryan Bednarz, PhD
Professor of Medical Physics
University of Wisconsin

Reinier Hernandez, PhD
Assistant Professor of Medical Physics
University of Wisconsin

Seungpyo Hong, PhD
Professor of Pharmaceutical Sciences
University of Wisconsin
Learn more:
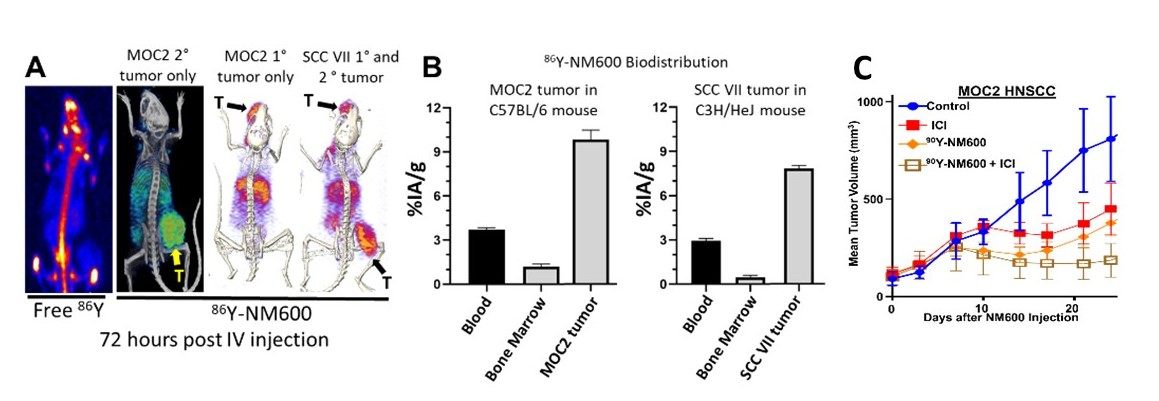
Figure. A. While IV injected 86Y localizes to bone marrow of mice on PET/CT (left), 86Y-NM600 shows selective uptake in 1° and 2° MOC2 and SCC VII tumors (T, indicated by arrow) in syngeneic mice at 72 hours post-IV injection. B. Measured biodistribution confirms selective uptake of NM600 in these HNSCC tumor models relative to blood and bone marrow. Preliminary dosimetry from these studies indicated that an injected activity of 50 µCi 90Y-NM600 will deliver 4 Gy to the tumor microenvironment for MOC2 and SCC VII tumors in syngeneic mice. C. In mice bearing MOC2 HNSCC flank tumors, we observe a cooperative effect of 50 µCi 90Y-NM600 and anti-CTLA-4 (ICI) resulting in improved tumor control.